This web page was produced as an assignment for Genetics 677, an undergraduate course at UW-Madison.
Nucleotide Sequence
The nucleotide sequence of the primary two isoforms of the GRIK2 gene localized on chromosome 6 were found using ENTREZ and GeneCards, which were ultimately directed to the NCBI website.
Accession Number: NG_009224
The FASTA formatted nucleotide sequence of isoform 1 (variant 1) of the GRIK2 sequence can be accessed here. This variant encodes the longer isoform (1).
The FASTA formatted nucleotide sequence of isoform 2 (variant 2) of the GRIK2 sequence can be accessed here. This variant contains an additional exon at the 3' end compared to transcript variant 1, causing a frame-shift and early translation termination. The resulting isoform is shorter and has a distinct C-terminus compared to isoform 1 (1).
A search of GRIK2 in the UniProt database yielded 5 isoforms of the GRIK2 protein shown below.
The GRIK2 gene (non-coding & coding) is 678026 base pairs, while the mRNA coding sequence is 4550 base pairs. Using either variant 1 or 2 given by Entrez provided the same results when using the databases cited on this website.
Isoforms
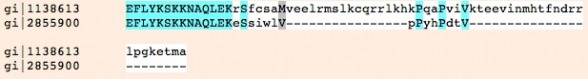
Shown to the left is a comparison of the two isoforms of GRIK2 using Muscle. As described above, isoform 2 (gi 2855900) is shorter with a distinct C-terminus.

The five isoforms found using the UniProt database.
Species Homologies
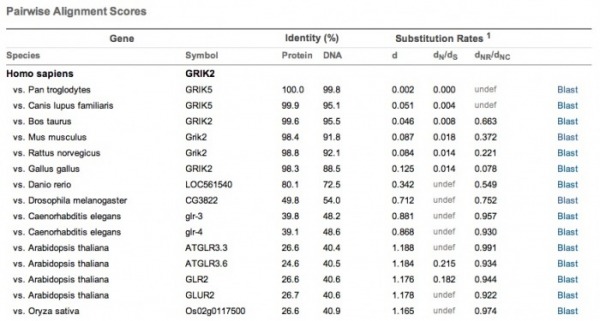
Click on the image to be directed to the HomoloGene results page for the pairwise alignments of GRIK2 and its putative homologues. As shown, the homologues demonstrate a high degree of similarity in their nucleotide sequences. Notably, the mouse (mus musculus), which is being used as a model organism for understanding GRIK2's role in bipolar disorder, has a very high degree of similarity in both DNA and protein sequence (91.8%/98.4%).
Multiple Alignment (DNA)
| tcoffeealignment.pdf | |
| File Size: | 114 kb |
| File Type: | |
The T-Coffee multiple alignment program was used to assess the amount of sequence similarity among the putative homologues in 12 species. The species used for the multiple alignment were the same ones given by HomoloGene in the above pairwise alignment. Click on the pdf link above to view the entire multiple alignment file. There are various sections of highly conserved sequence throughout the alignment, indicating possible locations for sequences that encode important functional parts of the protein. Given the function of GRIK2 as a glutamate receptor, these locations are likely to be involved in ligand binding.
Multiple Alignment - BLAST (protein)
| blastalignment.txt | |
| File Size: | 359 kb |
| File Type: | txt |
References:
1. NCBI Database. http://www.ncbi.nlm.nih.gov/sites/entrez?db=gene&cmd=Retrieve&dopt=full_report&list_uids=2898. Retrieved February 6, 2009.
BLAST
MUSCLE
T-COFFEE
HOMOLOGENE
GENECARDS
Ashley Bateman, [email protected], last updated 5/13/09
http://www.gen677.weebly.com
