This web page was produced as an assignment for Genetics 677, an undergraduate course at UW-Madison.
Gene Ontology
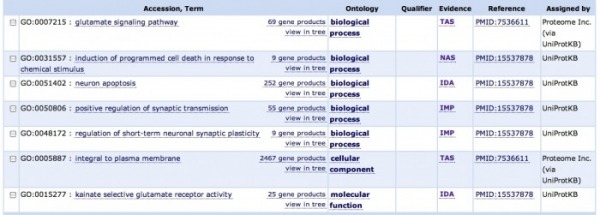
The image above is from GO Consortium, under the identification number of Q13002. There were 7 associations listed, with the majority of ontological classifications under biological process, along with one each under cellular component and molecular function. All of the accession terms make logical sense in the context of this gene's role as a glutamate kainate receptor. There are two obvious accession terms: glutamate signaling pathway and kainate selective glutamate receptor activity, the former under the biological process category and the latter under the molecular function category. 3 others represent a biological role in neuronal transmission events; The first two, positive regulation of synaptic transmission and regulation of short-term neuronal synaptic plasticity are under the biological process category, and the third, integral to plasma membrane, is under the cellular component category. The last two deal with cell death and are both under the biological process category: neuron apoptosis, and induction of programmed cell death in response to chemical stimulus. It is logical that as a receptor, it could be sensitive to stimuli that cause apoptosis, or programmed cell death. Other GO websites produced very similar output.
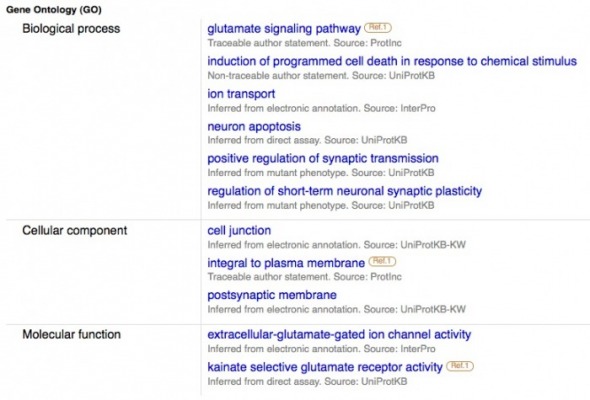
Gene ontology provided by Uniprot. Additional GO results are given, possibly because Uniprot samples different databases than the GO Consortium. The additional GO results are logical, however, given GRIK2's role as a glutamate receptor.
Phenotype: C. elegans
Using WormBase, glr-3 was found to be the most similar gene to GRIK2 in C. elegans. The WormBase gene id is WBGene00001614. Glr-3 encodes a protein that contains a ligand-gated ion channel domain and a receptor family ligand binding domain with similarity to kainate-selective ionotropic glutamate receptor 2 (GRIK2); expressed in the RIA neurons of C. elegans (shown above).
No phenotypes were observed in C. elegans in RNAi experiments.
Phenotype: D. melanogaster
Using FlyBase, Gene CG3822 was found to be the most similar gene to GRIK2 in D. melanogaster. The FlyBase gene id isFBgn0038837. Its molecular function is described as kainate selective glutamate receptor activity.
No phenotypes were observed in D. melanogaster in RNAi experiments.
It has been demonstrated that siRNA is not as effective at silencing mRNA expression in neuronal tissue. (1) Since the expression of this gene is in neuronal tissue, it is not surprising that these experiments yielded no resultant phenotype.
Other homologous organisms

No phenotypes from RNAi were found for any of the other model organisms listed in my phylogeny or homologene results. I did an extensive web and model organism database search, and no information other than basic genome location and sequence data could be found about the homologous genes.
This is unfortunate given the conserved nature of GRIK2 and its homologues. I am sure that valuable information about the functioning of GRIK2 and the role that inhibition of ligand binding plays in the resultant phenotype would be found using these organisms. Hopefully these experiments are currently being developed.
The image at left is of A. thaliana. (2).
Ashley Bateman, [email protected], last updated 5/13/09
http://www.gen677.weebly.com
References:
1. Sugimoto A. (2004). High-throughput RNAi in Caenorhabditis elegans: genome-wide screens and functional genomics. Differentiation. 2004 Mar;72(2-3):81-91. Review.
2. Arabidopsis image from http://www.botanik.uni-karlsruhe.de/garten/fotos-knoch/Arabidopsis%20thaliana%20%20Acker-Schmalwand%202%20_%2022%20Tage%20alt.jpg
Databases used:
GO Consortium
AMIGO
WormBase
FlyBase
RNAi Database