This web page was produced as an assignment for Genetics 677, an undergraduate course at UW-Madison.
Protein Data
Uniprot accession number: Q13002
Molecular weight/aa: 102,583 Da/908 aa
Theoretical Isoelectric Point: Theoretical pI/Mw: 8.05 / 102583.37 (http://us.expasy.org/cgi-bin/pi_tool)
For information concerning GRIK2's protein isoforms, see the Gene Sequences & Homology page.
For information concerning GRIK2's protein Gene Ontology, see the Phenotypes & Gene Ontology page.
Figure at left retrieved from String JMOL feature.
Protein Localization
Using a hybrid prediction approach based on sequence composition, physico-chemical properties, dipeptide composition, and psi-BLAST, at ESLpred, GRIK2 is predicted to be localized to the nucleus.
Based on the predicted presence of any of the N-terminal presequences, at TargetP, GRIK2 is predicted to contain a secretory pathway signal peptide (SP).
Based on the review of subcellular locations determined by a high-throughput, immunofluorescence-based assay and by manually reviewing peer-reviewed publications, at LOCATE, GRIK2 is predicted to be a secreted protein, a type I membrane protein, and a multipass membrane protein. Based on the occurrence patterns of protein functional domains and the amino acid compositional differences in proteins from different subcellular locations, at pTARGET, GRIK2 is predicted with 100% confidence to be localized to the plasma membrane.
Therefore, based on my knowledge of the function of GRIK2 as a kainate glutamate receptor along with the results from the above localization databases, I believe that GRIK2 is primarily localized to the plasma membrane, although it is possible that it could have other secondary functions that localize it to other regions in the cell, as described by the various localization algorithims above.
Protein Domains
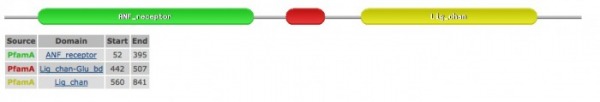
The above image and information below was retrieved through the PFAM database.
The ANF receptor shown in green is part of the family that includes includes extracellular ligand binding domains of a wide range of receptors, including GRIK2. This family also includes the bacterial amino acid binding proteins of known structure, as shown in the Phylogeny & Motifs page.
The domain shown in red is the luminal domain just upstream of the first transmembrane region ion-channel proteins (such as GRIK2) and it binds L-glutamate and glycine.
The domain shown in yellow is part of a family that includes the four transmembrane regions of the ionotropic glutamate receptors, such as GRIK2, as well as NMDA receptors.
Transmembrane Domains & Post-Translational Modification
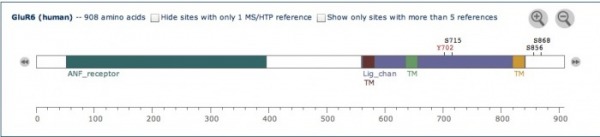
Using Phosphosite, I was able to locate 3 transmembrane domains, indicated by "TM" on the figure at left. In addition, four post-translational modification sites are shown: Y702, S715, S856, and S868.
According to Uniprot, a type of modification called sumoylation mediates kainate receptor-mediated endocytosis and regulates synaptic transmission of GRIK2.
In addition, glycosylation occurs throughout the protein sequence.
Trypsin Cleavage

Results found using ExPASy Peptide Cutter. Trypsin has 92 cleavage sites, at the positions indicated in the righthand column.
Databases used:
Uniprot
Pfam
ExPASy Peptide Cutter
Phosphosite
ESLpred
TargetP
LOCATE
pTARGET
Ashley Bateman, [email protected], last updated 4/26/09
http://www.gen677.weebly.com